COVER STORY SIDEBAR
Guy in the middle who picks apart roots, genes and software
Randy Clark '04 began photographing roots the year he graduated, when he began working for two years as a technician in the lab of Leon Kochian, director of the U.S. Department of Agriculture's Agricultural Research Service at Cornell.

Randy Clark '04, a graduate student in bioengineering, prepares the roots of a rice plant seedling for imaging in three dimensions. See larger image
Kochian, who studies aluminum toxicity in cereal crops, wanted to better understand how some plants withstand the metal's toxicity on acidic soils, so Clark grew tomato, sorghum and corn hydroponically, laid the plants' roots on a light table and photographed and measured them.
Five years later, Clark, now a Cornell graduate student in bioengineering, has devised a system of cylindrical glass containers and gel for growing rice seedlings and imaging their roots in three dimensions as they grow, and has also developed software to quantify details of each root's physical traits. Kochian's lab is part of a National Science Foundation-funded study of plant genome and genomics of root system architecture, that includes Duke and Georgia Tech. Kochian also collaborates with Cornell plant geneticist Susan McCouch to pick apart how a plant's genes relate to its root system architecture.
"Randy's a hybrid," says Kochian, who also has an adjunct appointment in the Department of Crop and Soil Sciences. "He can talk with the plant people, and he seamlessly talks with the computer scientists at Georgia Tech and Duke. He's written a lot of the software they're using now. I am really proud of him; we go to these meetings, and he's holding his own with the top people in the world."
"You need that guy in the middle who can see both viewpoints," says Clark. "I can cherry pick ideas and tools to use to move the project forward."
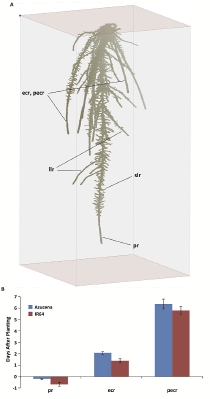
Clark's software can create 3-D images and animations that chart and predict root growth in rice plants based on specific genes. See larger image
Clark's system involves special glass cylinders that contain gellan gum, a bacterial byproduct that is dissolved in water with nutrients before it solidifies and serves as a growing medium. Clark and Janelle Jung, a graduate student in McCouch's lab, grow hundreds of rice seedlings, usually one per cylinder. On days three, six, nine and twelve of growth, a camera, synchronized with a rotating turntable, automatically takes 40 images of each root system, one image every 9 degrees, as the platform on which the cylinder sits rotates 360 degrees. The cylinder sits within a clear plastic box of water to eliminate refraction through the cylinder and to provide a clear image of the roots.
Eventually, thanks to Clark's software, the researchers end up with 3-D images and animations for each plant, which can be interactively rotated on a computer screen. The software also quantifies essential measurements and angles of the roots so that the data can eventually be used to link the nuances of root growth characteristics to specific genes or regions of each plant's genome.
Clark has a steady queue of plants to document: 400 cultivated rice varieties, 100 wild rice types, and then 170 offspring of a cross between a lowland rice variety bred for irrigated conditions with shallow roots and an upland dry-soil rice variety with a deep, searching tap root.
In an interesting discovery, Clark observed that roots don't grow straight down, but rather in a winding corkscrew pattern, like an inversion of climbing vines. The pattern has scant reference in the literature. "Maybe it has to do with mining the soil for nutrients and accessing more area," Clark speculates.
"We're looking at a portion of the plant no one has ever looked at in this way before," says McCouch.